Returns a vector of cell numbers at the locations of seed cells and growth buffers.
anchor.seed(
attTbl,
ngbList,
rNumb = FALSE,
class = NULL,
cond.filter = NULL,
cond.seed,
cond.growth = NULL,
lag.growth = Inf,
cond.isol = NULL,
lag.isol = 1,
sort.col = NULL,
sort.seed = "max",
saveRDS = NULL,
overWrite = FALSE,
isol.buff = FALSE,
silent = FALSE
)Arguments
- attTbl
data.frame, the attribute table returned by the function
attTbl.- ngbList
list, the list of neighborhoods returned by the function
ngbList.- rNumb
logic, the neighborhoods of the argument
ngbListare identified by cell numbers (rNumb=FALSE) or by row numbers (rNumb=TRUE) (seengbList). It is advised to use row numbers for large rasters.- class
numeric, the classification number to assign to all cells that meet the function conditions. If
NULL, a new class number is assigned every time a new seed cell is identified. Growth buffers have the same classification number as the seed cell to which they refer.- cond.filter
character string, defines for what cells the arguments
cond.seed,cond.growthandcond.isolhave to be evaluated. It can beNULL. Absolute conditions can be used (seeconditions).- cond.seed
character string, the conditions to identify seed cells. Absolute conditions can be used (see
conditions). It cannot beNULL.- cond.growth
character string, the conditions to define a growth buffer around seed cells. It can be
NULL. Absolute and focal cell conditions can be used (seeconditions).- lag.growth
0 or Inf, defines the evaluation lag of focal cell conditions in
cond.growth.- cond.isol
character string, the conditions to define an isolation buffer around seed cells and growth buffers. It can be
NULL. Absolute and focal cell conditions can be used (seeconditions).- lag.isol
0 or Inf, defines the evaluation lag of focal cell conditions in
cond.isol.- sort.col
character, the column name in the
attTblon which thesort.seedis based on. It determines in what order seed buffers are computed.- sort.seed
character, the order seed buffers are computed is based on the value seed cells have in the column of attribute table column named
sort.col. Ifsort.seed="max", buffers are computed from the seed cell having the maximum value to the seed cell having the minimum value. Ifsort.seed="min", buffers are computed in the opposite order.- saveRDS
filename, if a file name is provided save the class vector as an RDS file.
- overWrite
logic, if the RDS names already exist, existing files are overwritten.
- isol.buff
logic, return the isolation buffer (class = -999).
- silent
logic, progress is not printed on the console.
Value
Class vector. See conditions for more details about
class vectors.
Details
This function implements an algorithm to identify seed cells, growth
buffers and isolation buffers.
Condition arguments
The function takes as inputs four sets of conditions with
cond.growth and cond.isol taking into account class
contiguity and continuity (see conditions):
cond.filter, the conditions to define what cells have to be evaluated by the function.cond.seed, the conditions to identify, at each iteration, the seed cell. The seed cell is the cell around which growth and isolation conditions are applied.cond.growth, the conditions to define a buffer around the seed cell.cond.isol, the conditions to isolate one seed cell (and its growth buffer) from another.
Iterations
The argument cond.filter defines the set of cells to be considered
by the function.
A seed cell is identified based on
cond.seedand receives a classification number as specified by the argumentclass. Ifclass=NULL, then a new class is assigned to every new seed cell.Cells connected with the seed cell meeting the conditions of
cond.growthare assigned to the same class of the seed cell (growth buffer). The rule evaluation take into account class continuity (seeconditions).Cells connected with the seed cell (or with its growth buffer) meeting the conditions of
cond.isolare assigned to the isolation buffer (class = -999). The rule evaluation take into account class continuity (seeconditions).A new seed cell is identified based on
cond.seedwhich is now only evaluated for cells that were not identified as seed, growth or isolation cells in previous iterations.A new iteration starts. Seed, growth and isolation cells identified in previous iteration are ignored in successive iterations.
The function stops when it cannot identify any new seed cell.
Relative focal cell conditions and evaluation lag
The arguments
lag.growthandlag.isolcontrol the evaluation lag of relative focal cell conditions (seeconditions).When
lag.*are set to0, relative focal cell conditions have a standard behavior and compare the values of thetest cellsagainst the value of thefocal cell.When
lag.*are set toInf, relative focal cell conditions compare the values of thetest cellsagainst the value of theseed cellidentified at the start of the iteration.
See also
Examples
# DUMMY DATA
############################################################################
# LOAD LIBRARIES
library(scapesClassification)
library(terra)
# LOAD THE DUMMY RASTER
r <- list.files(system.file("extdata", package = "scapesClassification"),
pattern = "dummy_raster\\.tif", full.names = TRUE)
r <- terra::rast(r)
# COMPUTE THE ATTRIBUTE TABLE
at <- attTbl(r, "dummy_var")
# COMPUTE THE LIST OF NEIGBORHOODS
nbs <- ngbList(r)
############################################################################
# EXAMPLE PLOTS
############################################################################
oldpar <- par(mfrow = c(1,2))
m <- c(4.5, 0.5, 2, 3.2)
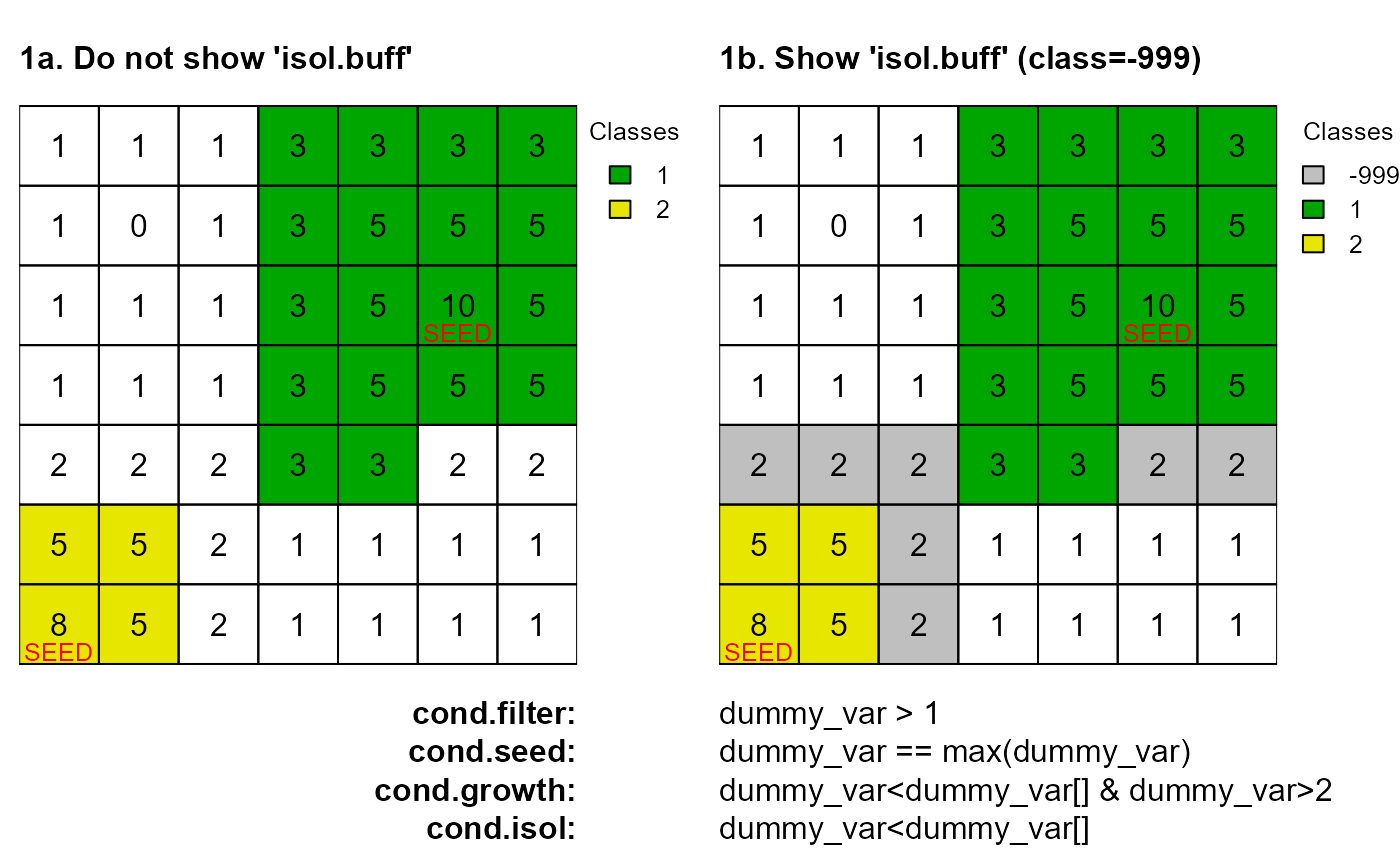
# 1a. Do not show isol.buff
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var == max(dummy_var)",
cond.growth = "dummy_var<dummy_var[] & dummy_var>2",
cond.isol = "dummy_var<dummy_var[]")
plot(cv.2.rast(r,classVector=as), type="classes", mar=m, col=c("#00A600", "#E6E600"),
axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "1a. Do not show 'isol.buff'")
mtext(side=1, line=0, cex=1, font=2, adj=1, "cond.filter:")
mtext(side=1, line=1, cex=1, font=2, adj=1, "cond.seed:")
mtext(side=1, line=2, cex=1, font=2, adj=1, "cond.growth:")
mtext(side=1, line=3, cex=1, font=2, adj=1, "cond.isol:")
text(xFromCell(r,c(20,43)),yFromCell(r,c(20,43))-0.05,"SEED",col="red",cex=0.80)
# 1b. Show isol.buff
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var == max(dummy_var)",
cond.growth = "dummy_var<dummy_var[] & dummy_var>2",
cond.isol = "dummy_var<dummy_var[]", isol.buff = TRUE)
plot(cv.2.rast(r,classVector=as), type="classes", col=c("#00000040", "#00A600", "#E6E600"),
mar=m, axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "1b. Show 'isol.buff' (class=-999)")
mtext(side=1, line=0, cex=1, adj=0, "dummy_var > 1")
mtext(side=1, line=1, cex=1, adj=0, "dummy_var == max(dummy_var)")
mtext(side=1, line=2, cex=1, adj=0, "dummy_var<dummy_var[] & dummy_var>2")
mtext(side=1, line=3, cex=1, adj=0, "dummy_var<dummy_var[]")
text(xFromCell(r,c(20,43)),yFromCell(r,c(20,43))-0.05,"SEED",col="red",cex=0.80)
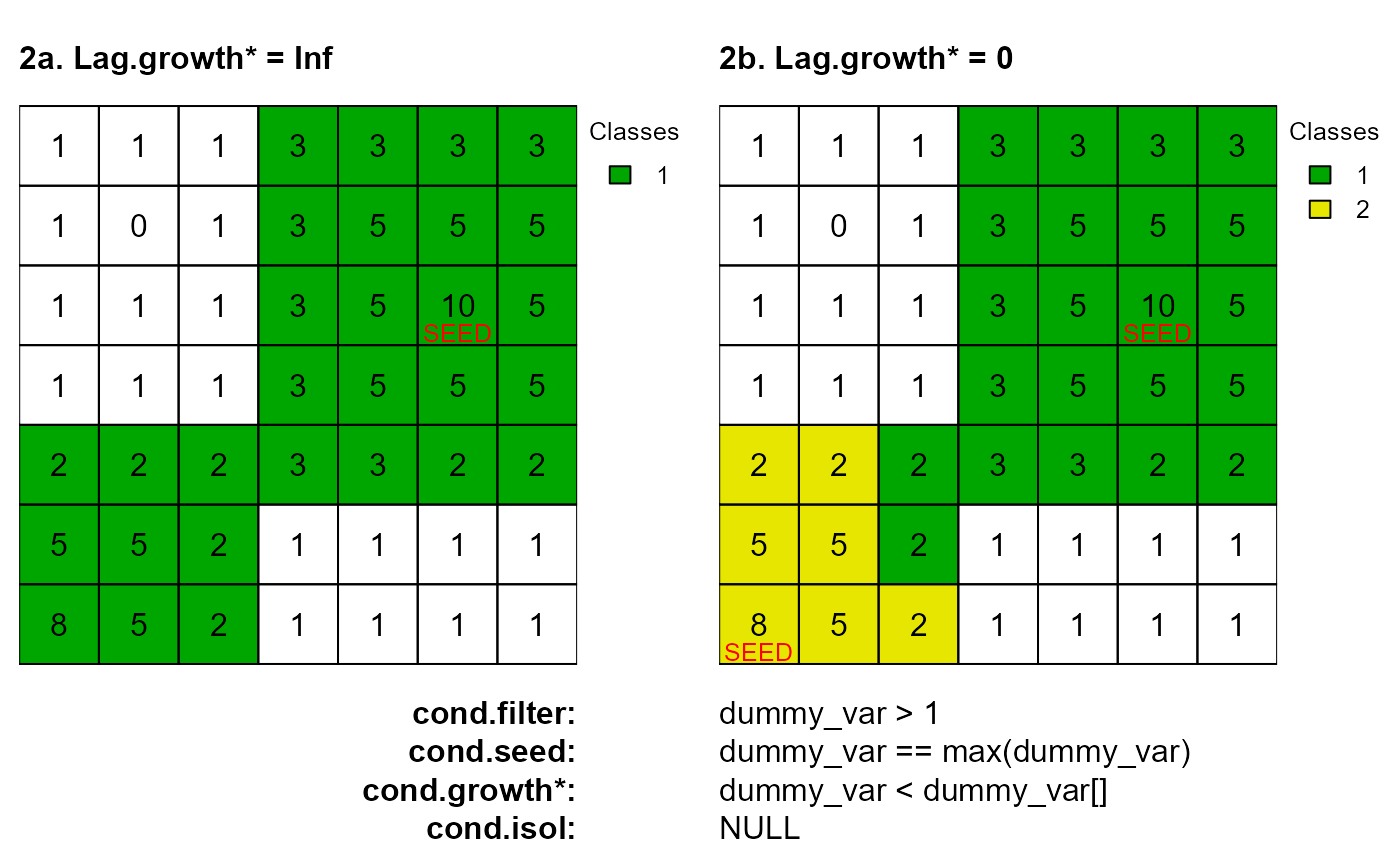
 # 2a. Lag.growth = Inf
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var == max(dummy_var)",
cond.growth = "dummy_var<dummy_var[]", lag.growth = Inf)
plot(cv.2.rast(r,classVector=as), type="classes", mar=m, col=c("#00A600"),
axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "2a. Lag.growth* = Inf")
mtext(side=1, line=0, cex=1, font=2, adj=1, "cond.filter:")
mtext(side=1, line=1, cex=1, font=2, adj=1, "cond.seed:")
mtext(side=1, line=2, cex=1, font=2, adj=1, "cond.growth*:")
mtext(side=1, line=3, cex=1, font=2, adj=1, "cond.isol:")
text(xFromCell(r,c(20)),yFromCell(r,c(20))-0.05,"SEED",col="red",cex=0.80)
# 2b. Lag.growth = 0
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var == max(dummy_var)",
cond.growth = "dummy_var<dummy_var[]", lag.growth = 0)
plot(cv.2.rast(r,classVector=as), type="classes", mar=m, col=c("#00A600", "#E6E600"),
axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "2b. Lag.growth* = 0")
mtext(side=1, line=0, cex=1, adj=0, "dummy_var > 1")
mtext(side=1, line=1, cex=1, adj=0, "dummy_var == max(dummy_var)")
mtext(side=1, line=2, cex=1, adj=0, "dummy_var < dummy_var[]")
mtext(side=1, line=3, cex=1, adj=0, "NULL")
text(xFromCell(r,c(20,43)),yFromCell(r,c(20,43))-0.05,"SEED",col="red",cex=0.80)
# 2a. Lag.growth = Inf
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var == max(dummy_var)",
cond.growth = "dummy_var<dummy_var[]", lag.growth = Inf)
plot(cv.2.rast(r,classVector=as), type="classes", mar=m, col=c("#00A600"),
axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "2a. Lag.growth* = Inf")
mtext(side=1, line=0, cex=1, font=2, adj=1, "cond.filter:")
mtext(side=1, line=1, cex=1, font=2, adj=1, "cond.seed:")
mtext(side=1, line=2, cex=1, font=2, adj=1, "cond.growth*:")
mtext(side=1, line=3, cex=1, font=2, adj=1, "cond.isol:")
text(xFromCell(r,c(20)),yFromCell(r,c(20))-0.05,"SEED",col="red",cex=0.80)
# 2b. Lag.growth = 0
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var == max(dummy_var)",
cond.growth = "dummy_var<dummy_var[]", lag.growth = 0)
plot(cv.2.rast(r,classVector=as), type="classes", mar=m, col=c("#00A600", "#E6E600"),
axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "2b. Lag.growth* = 0")
mtext(side=1, line=0, cex=1, adj=0, "dummy_var > 1")
mtext(side=1, line=1, cex=1, adj=0, "dummy_var == max(dummy_var)")
mtext(side=1, line=2, cex=1, adj=0, "dummy_var < dummy_var[]")
mtext(side=1, line=3, cex=1, adj=0, "NULL")
text(xFromCell(r,c(20,43)),yFromCell(r,c(20,43))-0.05,"SEED",col="red",cex=0.80)
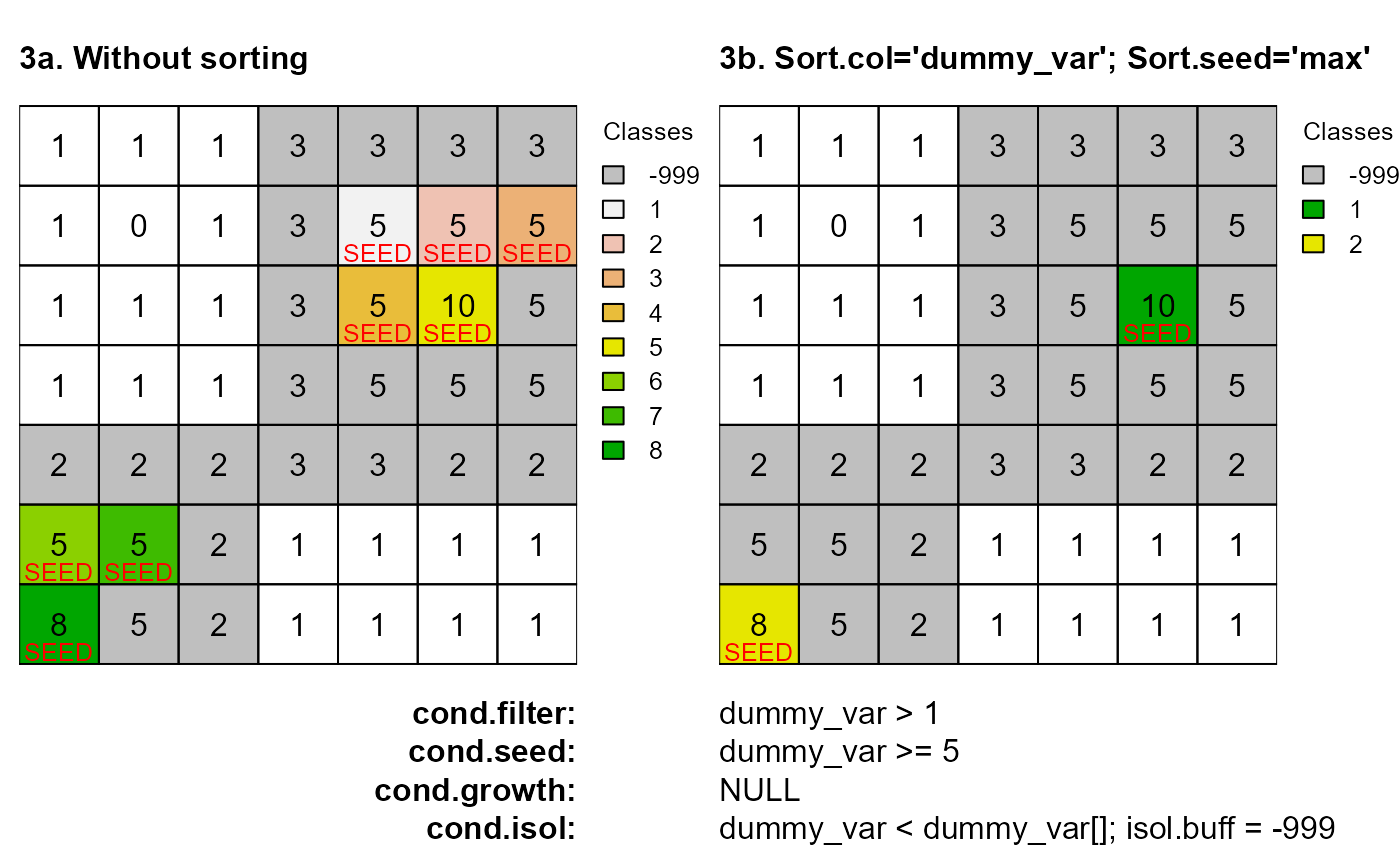
 # 3a. Without sorting
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var >= 5",
cond.isol = "dummy_var<dummy_var[]", isol.buff = TRUE)
seeds <- which(!is.na(as) & as !=-999)
cc <- c("#00000040", terrain.colors(8)[8:1])
plot(cv.2.rast(r,classVector=as), type="classes", mar=m, col=cc,
axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "3a. Without sorting")
mtext(side=1, line=0, cex=1, font=2, adj=1, "cond.filter:")
mtext(side=1, line=1, cex=1, font=2, adj=1, "cond.seed:")
mtext(side=1, line=2, cex=1, font=2, adj=1, "cond.growth:")
mtext(side=1, line=3, cex=1, font=2, adj=1, "cond.isol:")
text(xFromCell(r,seeds),yFromCell(r,seeds)-0.05,"SEED",col="red",cex=0.80)
# 3b. Sort buffer evaluation based on 'dummy_var' values
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var >= 5",
cond.isol = "dummy_var<dummy_var[]", isol.buff = TRUE,
sort.col = "dummy_var", sort.seed = "max")
seeds <- which(!is.na(as) & as !=-999)
plot(cv.2.rast(r,classVector=as), type="classes",col=c("#00000040", "#00A600", "#E6E600"),
mar=m, axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "3b. Sort.col='dummy_var'; Sort.seed='max'")
mtext(side=1, line=0, cex=1, adj=0, "dummy_var > 1")
mtext(side=1, line=1, cex=1, adj=0, "dummy_var >= 5")
mtext(side=1, line=2, cex=1, adj=0, "NULL")
mtext(side=1, line=3, cex=1, adj=0, "dummy_var < dummy_var[]; isol.buff = -999")
text(xFromCell(r,seeds),yFromCell(r,seeds)-0.05,"SEED",col="red",cex=0.80)
# 3a. Without sorting
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var >= 5",
cond.isol = "dummy_var<dummy_var[]", isol.buff = TRUE)
seeds <- which(!is.na(as) & as !=-999)
cc <- c("#00000040", terrain.colors(8)[8:1])
plot(cv.2.rast(r,classVector=as), type="classes", mar=m, col=cc,
axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "3a. Without sorting")
mtext(side=1, line=0, cex=1, font=2, adj=1, "cond.filter:")
mtext(side=1, line=1, cex=1, font=2, adj=1, "cond.seed:")
mtext(side=1, line=2, cex=1, font=2, adj=1, "cond.growth:")
mtext(side=1, line=3, cex=1, font=2, adj=1, "cond.isol:")
text(xFromCell(r,seeds),yFromCell(r,seeds)-0.05,"SEED",col="red",cex=0.80)
# 3b. Sort buffer evaluation based on 'dummy_var' values
as <- anchor.seed(attTbl = at, ngbList = nbs, rNumb = FALSE, class = NULL, silent = TRUE,
cond.filter = "dummy_var > 1", cond.seed = "dummy_var >= 5",
cond.isol = "dummy_var<dummy_var[]", isol.buff = TRUE,
sort.col = "dummy_var", sort.seed = "max")
seeds <- which(!is.na(as) & as !=-999)
plot(cv.2.rast(r,classVector=as), type="classes",col=c("#00000040", "#00A600", "#E6E600"),
mar=m, axes=FALSE, plg=list(x=1, y=1, cex=.80, title="Classes"))
text(r); lines(r)
mtext(side=3, line=0, cex=1, font=2, adj=0, "3b. Sort.col='dummy_var'; Sort.seed='max'")
mtext(side=1, line=0, cex=1, adj=0, "dummy_var > 1")
mtext(side=1, line=1, cex=1, adj=0, "dummy_var >= 5")
mtext(side=1, line=2, cex=1, adj=0, "NULL")
mtext(side=1, line=3, cex=1, adj=0, "dummy_var < dummy_var[]; isol.buff = -999")
text(xFromCell(r,seeds),yFromCell(r,seeds)-0.05,"SEED",col="red",cex=0.80)
 par(oldpar)
par(oldpar)