Check for spelling and syntax errors in conditions (cond argument) and
detect the type of conditions being used.
conditions(names_attTbl, cond, silent = FALSE)Arguments
- names_attTbl
character vector, the column (i.e. variable) names of the attribute table returned by the function
attTbl.- cond
character string, the condition string used by the
cond.*functions to classify raster cells (seeconditions).- silent
logic, when true, the function returns only error messages.
Value
An error message if the function finds spelling or syntax errors or a string with the types of rules detected in the condition string.
Details
Conditions (alias classification rules)
Classification rules evaluate either to true or false and determine what raster cells have to be classified.
Conditions are passed to
scapesClassificationfunctions as a single character string. They can consist of combination of variables names (as named in the attribute table, seeattTbl), arithmetic(+|-|*|/|^|%%|%/%), relational(>|<|>=|<=|==|!=|%/%)and logic operators(&||)and base R functions (e.g.,abs(variable_name)).All variables included in an attribute table (see
attTbl) can be included in a condition string by name (e.g., var name ="dummy_var"; condition ="dummy_var > 1").
Class vectors
Class vectors map raster cells to numeric classes.
The n^th^ element of a class vector stores the class of the raster cell stored in the n^th^ row of the corresponding attribute table (see
attTbl).Class vectors can serve also as a function input. As inputs, they provide information about the groups of cells that have already been classified.
Every time a class vector is provided as a function input, it is updated by assigning a numeric class to unclassified cells that meet function conditions.
Unclassified cells are represented as NA values.
Rule evaluation: Global evaluation
Classification rules are applied to all unclassified raster cells.
Function using global evaluation:
cond.4.all.
Rule evaluation: Focal evaluation
Classification rules are applied only to raster cells at specific locations and are based on class (dis)contiguity and class continuity.
Class contiguity:
classification rules are applied only to raster cells contiguous to focal cells (identified in thecond.*functions by the argumentnbs_of).Class continuity:
join into the same class cells that respect the same rules and that are connected to the same focal cells. This means that, at each iteration, newly classified cells become focal cells and conditions are tested in their neighborhood.Function using focal evaluation:
anchor.seed,cond.4.nofn,cond.reclass,reclass.nbsandclassify.all.
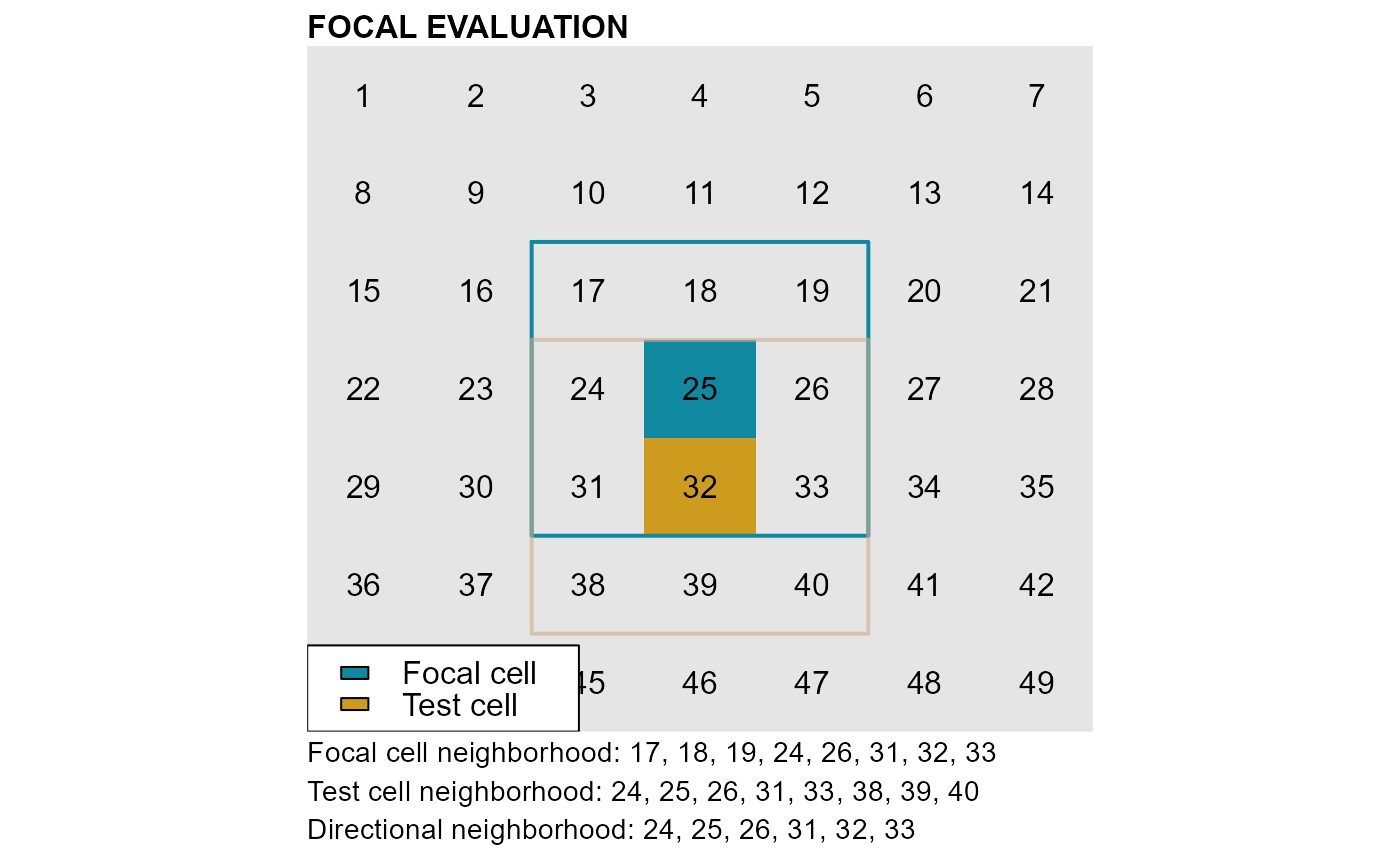
Focal evaluation: Definitions
Cell neighborhood: a cell with coordinates
(x, y)has 8 neighbors with coordinates:(x±1, y),(x, y±1)and(x±1, y±1). Cells on the edge of a raster have less than 8 neighbors. SeengbList.Focal cell: cells whose neighbors are evaluated against the classification rule(s). In the classification functions focal cells are identified by the argument
nbs_of.Test cell: the cell in the neighborhood of the focal cell that is being tested. At turns all cells in the neighborhood of a focal cell are tested against the classification rule(s).
Directional neighborhood: it consists of the intersection between the focal and the test cell neighborhoods.
Condition type: Absolute conditions
1) Absolute test cell condition: compares cell values against a threshold value.
This type of condition applies to all functions with a
conditionsargument.In global evaluations all cells meeting absolute conditions receive a classification number. In focal evaluations all
test cellsmeeting absolute conditions receive a classification number.Examples of valid conditions:
"variable_A > 1 & variable_B != 0";"(variable_A^2 < 50 & variable_B == 0) | abs(variable_C) > 50".
Functions:anchor.seed,cond.4.all,cond.4.nofnandcond.reclass.
2) Absolute neighborhood condition: compares the values of the
test cell and of its neighborhood against a threshold value.
An absolute neighborhood condition is identified by a variable name followed by curly brackets (e.g.,
"variable_name{}").A maximum of 9 evaluations are performed for each test cell (the test cell itself and the cells of its neighborhood are compared against a threshold value).
Test cells receive a classification number if the rule is true for at least as many evaluations as the ones specified by the argument
peval. The argumentpevalranges from 0 to 1. When 9 evaluations are performed,peval = 1means that all9evaluations have to be true;peval = 0.5means that at least4.5(rounded to5) evaluations have to be true.Only one neighborhood rule is allowed for each condition string (e.g., it is not possible to have a condition string like
"variable_A{} > 0 & variable_B{} > 1").The function
cond.4.nofncan consider adirectional neighborhoodinstead of the test cell neighborhood by setting the argumentdirectional = TRUE.Example of valid conditions:
"variable_A{} > 1 & abs(variable_B) != 0".
Functions:cond.4.nofnandcond.reclass.
Condition type: Relative conditions
1) Relative focal cell condition: compares the test cell value
against the focal cell value.
A relative focal cell condition is identified by a variable name followed by square brackets (e.g.,
"variable_name[]").Rules are defined repeating twice the same variable name, once with square brackets and once without. Square brackets indicate the focal cell value. As an example, the rule
"dummy_var < dummy_var[]"compares the value of the the test cell ("dummy_var") against the value of the focal cell ("dummy_var[]").Test cells are classified if the rule is true.
Examples of valid conditions:
"variable_A > variable_A[]";"(variable_A > variable_A[] & variable_B{} < 10) | variable_C > 1". Note that the last example is a combination of absolute and focal cell conditions.
Functions:anchor.seedandcond.4.nofn.
2) Relative neighborhood condition: compares the values of the test
cell against the values of the test cell neighborhood.
A relative neighborhood condition is identified by a variable name followed by curly brackets (e.g.,
"variable_name{}").Rules are defined repeating twice the same variable name, once with curly brackets and once without. Curly brackets indicate the test cell neighborhood. As an example, the rule
'dummy_var < dummy_var{}'compares the value of the the test cell (dummy_var) against the values of cells included in the test cell neighborhood (dummy_var{}).A maximum of 8 evaluations are performed for each test cell (the test cell is compared against each cell included in its neighborhood).
Test cells receive a classification number if the rule is true for at least as many evaluations as the ones specified by the argument
peval. The argumentpevalranges from 0 to 1. When 8 evaluations are performed,peval = 1means that all8evaluations have to be true;peval = 0.5means that at least4evaluations have to be true.Only one neighborhood rule is allowed for each condition string (e.g., it is not possible to have a condition string like
"variable_A{} > 0 & variable_B{} > variable_B").The function
cond.4.nofncan consider adirectional neighborhoodinstead of the test cell neighborhood by setting the argumentdirectional = TRUE.Example of valid conditions:
"variable_A > variable_A{}";"(variable_A > variable_A{} & variable_B != variable_B[]) | variable_C > 1". Note that the last example is a combination of absolute and relative conditions.
Functions:cond.4.nofnandcond.reclass.
See also
Examples
# LOAD LIBRARIES
library(scapesClassification)
library(terra)
################################################################################
# TYPES OF CONDITIONS
################################################################################
# As an example consider an attribute with the following columns
names_attTbl <- c("bathymetry", "slope")
# And the following conditions
cond <- "bathymetry>10"
conditions(names_attTbl, cond)
#> [1] "'Absolute test cell' condition type(s) detected."
cond <- "bathymetry[]>bathymetry | abs(slope{}) < 5"
conditions(names_attTbl, cond)
#> [1] "'Relative focal cell' AND 'Absolute neighborhood' condition type(s) detected."
cond <- "bathymetry[]>bathymetry | abs(slope{}) < slope"
conditions(names_attTbl, cond)
#> [1] "'Relative focal cell' AND 'Relative neighborhood' condition type(s) detected."
################################################################################
# FOCAL EVALUATION DEFINITIONS
################################################################################
# CELL NUMBERS OF A DUMMY RASTER (7X7)
r <- terra::rast(matrix(1:49, nrow = 7, byrow = TRUE), extent=c(0,7,0,7))
nbs <- ngbList(r)
# CLASS VECTOR WITH ONE TEST AND ONE FOCAL CELL
cv <- as.numeric(rep(NA, 49))
cv[c(32, 25)] <- c(1, 2) # tc (class 1), fc (class 2)
r_cv <- cv.2.rast(r, classVector = cv)
# POLYGONS REPRESENTING NEIGHBORHOODS
fcn <- rbind(c(2,5), c(5,5), c(5,2), c(2,2), c(2,5))
fcn <- terra::vect(fcn, type="polygons")
tcn <- rbind(c(2,4), c(5,4), c(5,1), c(2,1), c(2,4))
tcn <- terra::vect(tcn, type="polygons")
# PLOT - FOCAL EVALUATION DEFINITIONS
m <- c(3.5, 8, 1.2, 8)
plot(r_cv, type = "class", asp = NA, legend = FALSE, axes = FALSE, mar = m,
col = c("goldenrod3","#1088a0"), colNA= "grey90")
text(r)
mtext(side=3, line=0, adj=0, cex=1, font=2, "FOCAL EVALUATION")
mtext(side=1, line=0, adj=0, cex=0.9,
"Focal cell neighborhood: 17, 18, 19, 24, 26, 31, 32, 33")
mtext(side=1, line=1, cex=0.9, adj=0,
"Test cell neighborhood: 24, 25, 26, 31, 33, 38, 39, 40")
mtext(side=1, line=2, cex=0.9, adj=0,
"Directional neighborhood: 24, 25, 26, 31, 32, 33")
lines(fcn, col="#1088a0", lwd=2)
lines(tcn, col="#cfad8999", lwd=2)
legend("bottomleft", ncol = 1, bg = "white", y.intersp= 1.3,
legend = c("Focal cell", "Test cell"), fill = c("#1088a0", "goldenrod3"))