Classify all cells in classVector that have not yet been classified
based on contiguity and continuity conditions.
classify.all(attTbl, ngbList, rNumb = FALSE, classVector)Arguments
- attTbl
data.frame, the attribute table returned by the function
attTbl.- ngbList
list, the list of neighborhoods returned by the function
ngbList.- rNumb
logic, the neighborhoods of the argument
ngbListare identified by cell numbers (rNumb=FALSE) or by row numbers (rNumb=TRUE) (seengbList). It is advised to use row numbers for large rasters.- classVector
numeric vector, defines the cells in the attribute table that have already been classified. See
conditionsfor more information about class vectors.
Value
Update classVector with the new cells that were classified by
the function. See conditions for more details about class
vectors.
Details
The neighborhood of unclassified cells is considered. Among
neighbors, the class with the highest number of members is assigned to the
unclassified cell. If two or more classes have the same number of members,
then one of these classes is assigned randomly to the unclassified cell.
The function considers class continuity, thus, even cells that at first
were not contiguous to any class will be classified if continuous with at
least one cell having a class (see conditions).
See also
Examples
# DUMMY DATA
############################################################################
# LOAD LIBRARIES
library(scapesClassification)
library(terra)
# LOAD THE DUMMY RASTER
r <- list.files(system.file("extdata", package = "scapesClassification"),
pattern = "dummy_raster\\.tif", full.names = TRUE)
r <- terra::rast(r)
# COMPUTE THE ATTRIBUTE TABLE
at <- attTbl(r, "dummy_var")
# COMPUTE THE LIST OF NEIGBORHOODS
nbs <- ngbList(r)
################################################################################
# CLASSIFY.ALL
################################################################################
# compute example class vector
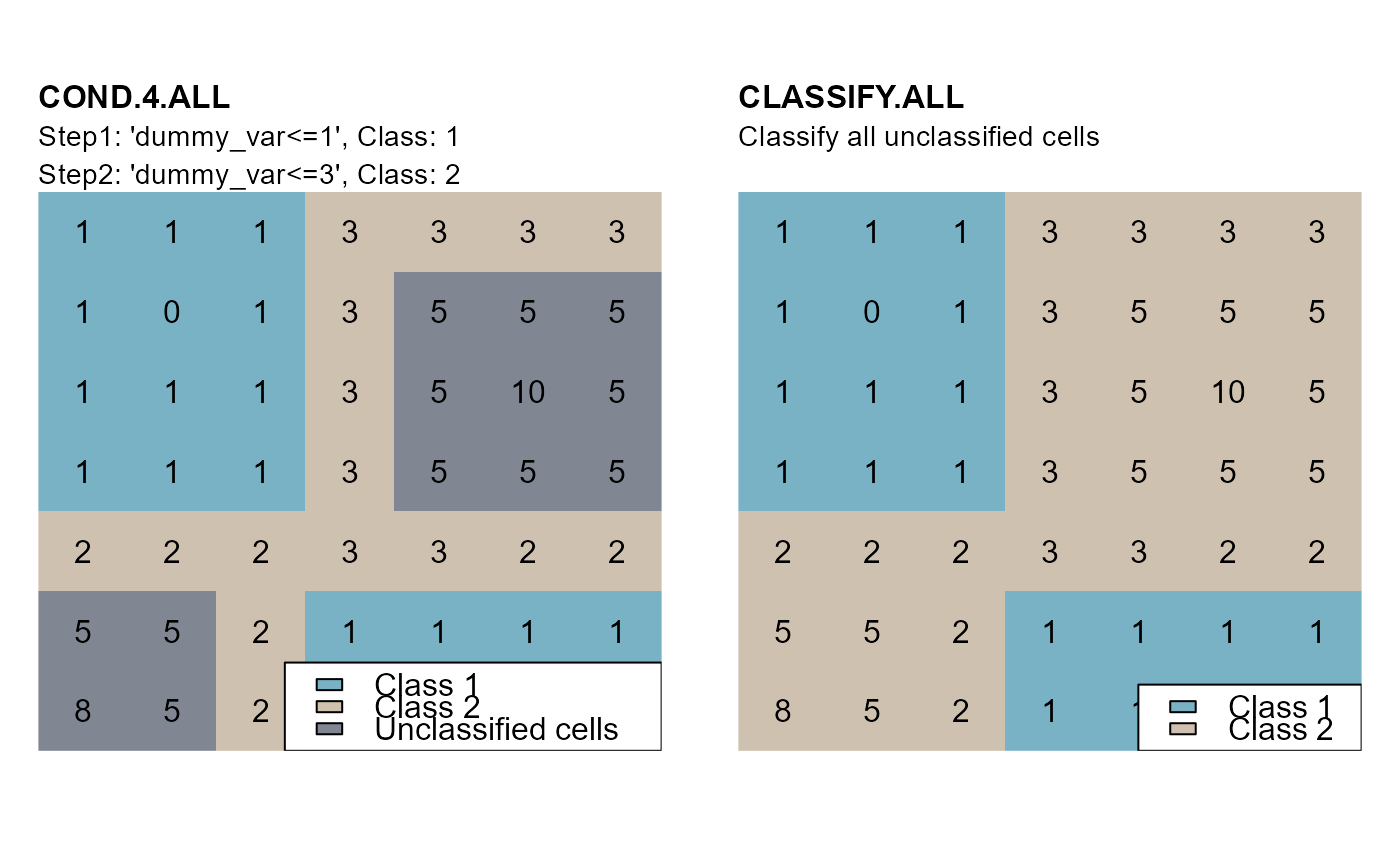
cv <- cond.4.all(attTbl = at, cond = "dummy_var <= 1", class = 1)
# update example calss vector
cv <- cond.4.all(attTbl = at, cond = "dummy_var <= 3", class = 2,
classVector = cv) # input previous class vector
# classify all unclassified cells
ca <- classify.all(attTbl = at, ngbList = nbs, rNumb = TRUE, classVector = cv)
# Convert class vectors into rasters
r_cv <- cv.2.rast(r, at$Cell, classVector = cv)
r_ca <- cv.2.rast(r, at$Cell,classVector = ca)
################################################################################
# PLOTS
################################################################################
oldpar <- par(mfrow = c(1,2))
m <- c(3, 1, 5, 1)
# 1)
plot(r_cv, type="classes", axes=FALSE, legend=FALSE, asp=NA, mar=m,
colNA="#818792", col=c("#78b2c4", "#cfc1af"))
text(r)
mtext(side=3, line=2, adj=0, cex=1, font=2, "COND.4.ALL")
mtext(side=3, line=1, adj=0, cex=0.9, "Step1: 'dummy_var<=1', Class: 1")
mtext(side=3, line=0, adj=0, cex=0.9, "Step2: 'dummy_var<=3', Class: 2")
legend("bottomright", bg = "white", fill = c("#78b2c4", "#cfc1af", "#818792"),
legend = c("Class 1", "Class 2", "Unclassified cells"))
# 2)
plot(r_ca, type="classes", axes=FALSE, legend=FALSE, asp=NA, mar=m,
colNA="#818792", col=c("#78b2c4", "#cfc1af"))
text(r)
mtext(side=3, line=2, adj=0, cex=1, font=2, "CLASSIFY.ALL")
mtext(side=3, line=1, adj=0, cex=0.9, "Classify all unclassified cells")
legend("bottomright", bg = "white", fill = c("#78b2c4", "#cfc1af", "#818792"),
legend = c("Class 1", "Class 2"))
 par(oldpar)
par(oldpar)