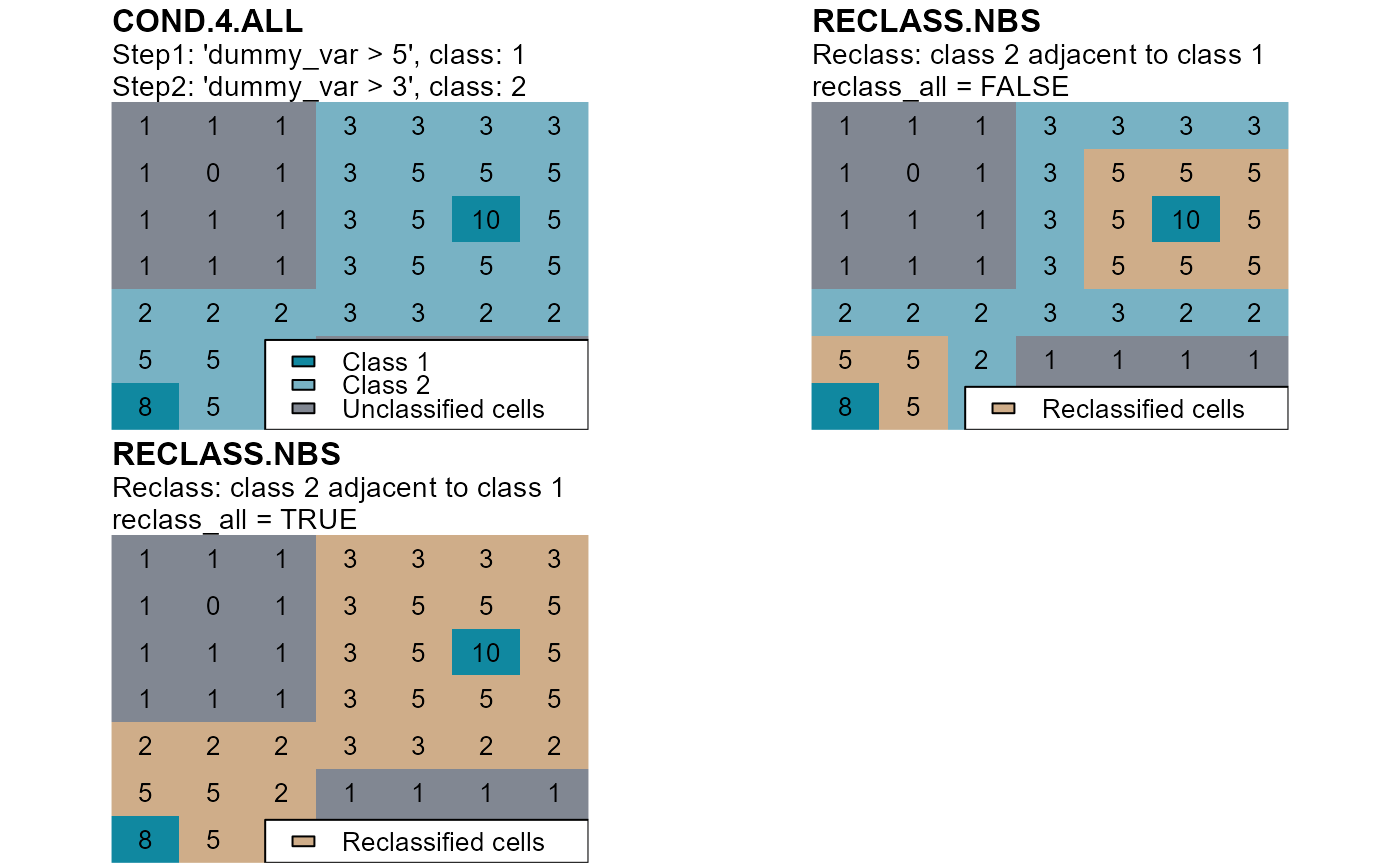
Evaluate if members of two classes are contiguous and, if they are, one of them is reclassified.
reclass.nbs(
attTbl,
ngbList,
rNumb = FALSE,
classVector,
nbs_of,
class,
reclass,
reclass_all = TRUE
)Arguments
- attTbl
data.frame, the attribute table returned by the function
attTbl.- ngbList
list, the list of neighborhoods returned by the function
ngbList.- rNumb
logic, the neighborhoods of the argument
ngbListare identified by cell numbers (rNumb=FALSE) or by row numbers (rNumb=TRUE) (seengbList). It is advised to use row numbers for large rasters.- classVector
numeric vector, defines the cells in the attribute table that have already been classified. See
conditionsfor more information about class vectors.- nbs_of
numeric or numeric vector, indicates the class(es) of focal and anchor cells.
- class
numeric or numeric vector, cells of classes
classadjacent to cells belonging to one of the classes ofnbs_ofare reclassified as indicated by the argumentreclass.- reclass
numeric, the classification number to assign to all cells that meet the function conditions.
- reclass_all
logic, all cells of class
classare also reclassified if they are connected to a reclassified cell.
Value
Update classVector with the new cells that were classified by
the function. See conditions for more information about class
vectors.
Details
The function evaluates if a cell of class
classis adjacent to a cell of classnbs_ofand, if it is, it is reclassifies as indicated by the argumentreclass.If the argument
reclass_all = TRUE, all cells of classclassare also reclassified if they are connected to a reclassified cell.
See also
Examples
library(scapesClassification)
library(terra)
# LOAD THE DUMMY RASTER
r <- list.files(system.file("extdata", package = "scapesClassification"),
pattern = "dummy_raster\\.tif", full.names = TRUE)
r <- terra::rast(r)
# COMPUTE THE ATTRIBUTE TABLE
at <- attTbl(r, "dummy_var")
# COMPUTE THE LIST OF NEIGBORHOODS
nbs <- ngbList(r)
################################################################################
# RECLASS.NBS
################################################################################
# Compute an inital class vector with `cond.4.all`
cv <- cond.4.all(attTbl = at, cond = "dummy_var > 5", class = 1)
# Update the class vector with a second class
cv <- cond.4.all(attTbl = at, cond = "dummy_var >= 2", class = 2,
classVector = cv)
# Reclassify cells of class 2 adjacent to cells of class 1
# reclass_all = FALSE
rc1 <- reclass.nbs(attTbl = at, ngbList = nbs,
# CLASS VECTOR `cv`
classVector = cv,
# CELLS OF CLASS...
class = 2,
# ...ADJACENT TO CELLS OF ANOTHER CLASS...
nbs_of = 1,
# ...WILL BE RECLASSIFIED...
reclass = 3,
# NO MORE RECLASSIFICATIONS
reclass_all = FALSE)
# reclass_all = TRUE
rc2 <- reclass.nbs(attTbl = at, ngbList = nbs,
# CLASS VECTOR `cv`
classVector = cv,
# CELLS OF CLASS...
class = 2,
# ...ADJACENT TO CELLS OF ANOTHER CLASS...
nbs_of = 1,
# ...WILL BE RECLASSIFIED...
reclass = 3,
# ...AND SO ALL CELLS OF CLASS 1 CONNECTED TO A RECLASSIFIED CELL
reclass_all = TRUE)
# Convert class vectors to rasters
r_cv <- cv.2.rast(r, at$Cell,classVector = cv, plot = FALSE)
r_rc1 <- cv.2.rast(r, at$Cell,classVector = rc1, plot = FALSE)
r_rc2 <- cv.2.rast(r, at$Cell,classVector = rc2, plot = FALSE)
################################################################################
# PLOTS
################################################################################
oldpar <- par(mfrow = c(2,2))
m = c(0.1, 3.5, 3.2, 3.5)
# 1)
plot(r_cv, type="classes", axes=FALSE, legend = FALSE, asp = NA, mar=m,
colNA="#818792", col=c("#1088a0", "#78b2c4"))
text(r)
mtext(side=3, line=2, adj=0, cex=1, font=2, "COND.4.ALL")
mtext(side=3, line=1, adj=0, cex=0.9, "Step1: 'dummy_var > 5', class: 1")
mtext(side=3, line=0, adj=0, cex=0.9, "Step2: 'dummy_var > 3', class: 2")
legend("bottomright", ncol = 1, bg = "white", y.intersp= 1.2,
legend = c("Class 1", "Class 2", "Unclassified cells"),
fill = c("#1088a0", "#78b2c4", "#818792"))
# 2)
plot(r_rc1, type="classes", axes=FALSE, legend = FALSE, asp = NA, mar=m,
colNA="#818792", col=c("#1088a0", "#78b2c4", "#cfad89"))
text(r)
mtext(side=3, line=2, adj=0, cex=1, font=2, "RECLASS.NBS")
mtext(side=3, line=1, adj=0, cex=0.9, "Reclass: class 2 adjacent to class 1")
mtext(side=3, line=0, adj=0, cex=0.9, "reclass_all = FALSE")
legend("bottomright", ncol = 1, bg = "white", y.intersp= 1.2,
legend = c("Reclassified cells"), fill = c("#cfad89"))
# 3)
plot(r_rc2, type="classes", axes=FALSE, legend = FALSE, asp = NA, mar=m,
colNA="#818792", col=c("#1088a0", "#78b2c4", "#cfad89"))
text(r)
mtext(side=3, line=2, adj=0, cex=1, font=2, "RECLASS.NBS")
mtext(side=3, line=1, adj=0, cex=0.9, "Reclass: class 2 adjacent to class 1")
mtext(side=3, line=0, adj=0, cex=0.9, "reclass_all = TRUE")
legend("bottomright", ncol = 1, bg = "white", y.intersp= 1.2,
legend = c("Reclassified cells"), fill = c("#cfad89"))
par(oldpar)